Evaluating Rosify Difodo VS SD-SLAM
As the previous entry concluded, the next step is to evaluate the performance of both odometry/slam algorithms, rosify_difodo and SD-SLAM.
Processing TUM dataset
In order to make this task easier and allow easy reproducibility, Ive created automated scripts to process with one script all TUM sequences in rosbag format within a directory. For this two scripts has been created:
- process_one_TUM_sequence.sh
- process_all_sequences_TUM.sh
Both scripts can be found on the scripts repo directory: https://github.com/RoboticsLabURJC/2019-tfm-omar-garrido/tree/master/scripts
process_one_TUM_sequence.sh
This script receives as an argument a sequence or rosbag from then TUM dataset. Then it creates a groundturuth file for SD-SLAM and rosify_difodo. The output directory where result files for each algorithm are going to be created are within the configuration of the odometry_evaluation_creator_file rospackage.
How to use
$ process_one_TUM_sequence.sh myrosbagfilefromTUM.rosbag
process_all_sequences_TUM.sh
This script gets as an argument a directory where rosbag sequences from TUM dataset are located within. All the sequences within this directory will be processed to create the results of odometry for SD-SLAM and rosify_difodo by executing process_one_TUM_sequence.sh.
How to use
./process_all_sequences_TUM.sh /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/myrosbagSequences/
Order of files created
Once it has been finished, one can do the following to know the order that the sequences have been generated, in order to change the name so they have meanignful names.
This will get the order of the files that has been processed.
$ cat processed_files_order.log
Proccessing file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_long_office_household.bag on sáb feb 22 12:18:24 CET 2020
Proccessing file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_notexture_far.bag on sáb feb 22 12:23:25 CET 2020
Proccessing file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_notexture_near_withloop.bag on sáb feb 22 12:24:18 CET 2020
Proccessing file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_texture_far.bag on sáb feb 22 12:26:17 CET 2020
Proccessing file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_texture_near_withloop.bag on sáb feb 22 12:27:08 CET 2020
Proccessing file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_halfsphere.bag on sáb feb 22 12:30:01 CET 2020
Proccessing file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_rpy.bag on sáb feb 22 12:32:09 CET 2020
Proccessing file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_static.bag on sáb feb 22 12:33:46 CET 2020
Proccessing file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_xyz.bag on sáb feb 22 12:35:11 CET 2020
Proccessing file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_structure_notexture_far.bag on sáb feb 22 12:37:41 CET 2020
To get the exact time where rosify_difodo output file was created for each file do:
$ cat process_all_debug.log | grep ROSIFY_DIFODO
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_long_office_household.bag on sáb feb 22 12:18:24 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_notexture_far.bag on sáb feb 22 12:23:25 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_notexture_near_withloop.bag on sáb feb 22 12:24:18 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_texture_far.bag on sáb feb 22 12:26:17 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_texture_near_withloop.bag on sáb feb 22 12:27:08 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_halfsphere.bag on sáb feb 22 12:30:01 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_rpy.bag on sáb feb 22 12:32:09 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_static.bag on sáb feb 22 12:33:46 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_xyz.bag on sáb feb 22 12:35:11 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_structure_notexture_far.bag on sáb feb 22 12:37:41 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_structure_notexture_near.bag on sáb feb 22 12:39:08 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_structure_texture_far.bag on sáb feb 22 12:41:01 CET 2020-------------
-------------ROSIFY_DIFODO: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_structure_texture_near.bag on sáb feb 22 12:42:51 CET 2020-------------
To get the exact time where SD-SLAM output file was created for each file do:
$ cat process_all_debug.log | grep "SD-SLAM RGBD"
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_long_office_household.bag on sáb feb 22 12:21:55 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_notexture_far.bag on sáb feb 22 12:23:59 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_notexture_near_withloop.bag on sáb feb 22 12:25:36 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_texture_far.bag on sáb feb 22 12:26:49 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_nostructure_texture_near_withloop.bag on sáb feb 22 12:29:01 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_halfsphere.bag on sáb feb 22 12:31:28 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_rpy.bag on sáb feb 22 12:33:15 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_static.bag on sáb feb 22 12:34:44 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_sitting_xyz.bag on sáb feb 22 12:36:56 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_structure_notexture_far.bag on sáb feb 22 12:38:37 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_structure_notexture_near.bag on sáb feb 22 12:40:21 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_structure_texture_far.bag on sáb feb 22 12:42:16 CET 2020-------------
-------------SD-SLAM RGBD: Process file: /home/omar/Documents/rosbag/rgbd_TUM/freidburg3/sequences//rgbd_dataset_freiburg3_structure_texture_near.bag on sáb feb 22 12:44:15 CET 2020-------------
Comparing results with groundtruth
The tool thats is going to be use to compare the results of SD-SLAM and rosify_difodo against the groundtruthof TUM dataset is EVO
How to install and use EVO was already discuss in a previous entry on this blog, check it here.
In order to do this Ive created a python script that allows to compare using evo package as an “import”.
compare_groundtruth_odometry_TUM.py
This python script that can be found here allows to compare multiple groundtruth files within a directory against the estimated files from another directory. Both directories are passed as arguments to the python script. The script expects the estimated files to have the same “name of sequence” as the groundtruth in order to find them. For example: Groundtruth file from TUM: rgbd_dataset_freiburg1_desk-groundtruth.txt Expected name for estimated file: *f1_desk-*.txt where “*” can be anything. f1 means that is a freiburg1 sequence and then desk is the name of the original sequence. The resulst are two files:
- ape_results.csv
- rpe_results.csv
On each file the results for each trajectory are presented.
Results
Using TUM RGBD dataset with RPE and ATE
Since Im going to use the TUM dataset to validate against, Ill use their original paper to see how should I measure the algorithms with their dataset. The paper presents two metrics for the evaluation of odometry and SLAM algorithms:
- RPE
- ATE or APE
RPE
RPE stands for Relative Pose Error. From the paper we get that the RPE measures the local accuracy of a trajectory over a fixed time interval ∆. Therefore, the relative pose error corresponds to the drift of the trajectory which is in particular useful for the evaluation of visual odometry systems. The suggestion on the paper is to use the RPE metric, since thats what a lot of papers have been using to compare their algorithms. But RPE can be used in different ways by changing the “delta” value that represents a fixed time interval (can also be frames). Also, and I cite: “Furthermore, the time parameter ∆ needs to be chosen. For visual odometry systems that match consecutive frames, ∆=1 is an intuitive choice”
So it seems, that if your odometry systems gets odometry from consecutive frames you should use 1 frame as delta value. Well this is what Ive done with evo and the results seems better than they are in reality. While the RPE seems to be in a range of > 10cm most os the times, the ATE can go up to more than a metter in some cases, so in my particular test I cannot trust this metric, RPE, to be a good measurement since a bad trajectory can give a good RPE. For example there is a case where SD-SLAM couldnt be initialize so all the points from the odometry where in the starting point but still the RPE value that I get was a good value of a few centimeters error.
Since RPE just look the differences in pose (translation/rotation) between frames and we have 30FPS and a speed which is not really big the movements between frames are small (just a few cm), the RPE wont be big with this approach even if the estimation is of the pose is null all the time.
A recommendation from the paper that will probably solve this problem, is to get the RPE for all the possibles ∆ values from 1 (to compare consecutive frames) to ∆=n (where n stands for the number of frames from out trajectory) and then average all values. This can be too expensive computationally speaking and also increases with the trajectory length. For this reasons I prefer to use ATE in this case to compare the results since it seems much easier.
ATE
ATE stands for Absolute Trajectory Errror. ATE on the other hand, does compute the global consistency of the estimated trajectory. This requires that the trajectories are aligned if needed. In my case I didnt aligned them with Uleyamas algoritm from evo since the estimated odometry results were already in the same frame than the original data, thanks to odometry_evalaution_file_creator ros package. ATE seems to be a much better measure in my case even though it only uses the translational error. The rotation error is also in the ATE in an inderect way since in the end a bad rotation translates to a bad pose or translation.
For the purpose of comparison Ill use the ATE due to the reasons I previously stated.
Once I compute all the results for several datasets from freiburg this are the results that I obtained for each algorithm.
Freiburg 1 sequences
| ATE: Rosify Difodo freiburg 1 | |||||||
|---|---|---|---|---|---|---|---|
| sequence_name | rmse | mean | median | std | min | max | sse |
| rgbd_dataset_freiburg1_desk2-groundtruth.txt | 0.262036876267081 | 0.24960420915859344 | 0.24517361048561637 | 0.0797562743244861 | 0.03932200401810672 | 0.3800444579256485 | 43.25789445 |
| rgbd_dataset_freiburg1_floor-groundtruth.txt | 0.8685882813484658 | 0.7244307368101799 | 0.702454899818355 | 0.4792136371813111 | 0.00774402995861971 | 1.6450140303353038 | 915.89696143 |
| rgbd_dataset_freiburg1_360-groundtruth.txt | 0.45069269054562916 | 0.42933429024562975 | 0.45185310666188855 | 0.1370983899633386 | 0.03917001404135568 | 0.6404594210408651 | 153.35854548999998 |
| rgbd_dataset_freiburg1_rpy-groundtruth.txt | 0.07933020605153356 | 0.0730068880566441 | 0.07219432109779872 | 0.031036686170778722 | 0.006116371473349228 | 0.14167843872657562 | 4.50598962 |
| rgbd_dataset_freiburg1_xyz-groundtruth.txt | 0.08006233836928092 | 0.07540128108124929 | 0.06690934164972787 | 0.026918856522216397 | 0.034418890162234934 | 0.1381616444603928 | 5.09593253 |
| rgbd_dataset_freiburg1_desk-groundtruth.txt | 0.1590195736316124 | 0.147940665685963 | 0.14208181941350673 | 0.05831624331499696 | 0.011270314991161568 | 0.29304206182730835 | 15.02061153 |
| rgbd_dataset_freiburg1_room-groundtruth.txt | 0.4148573774801919 | 0.3839373431062268 | 0.38442077207143743 | 0.15715839213441318 | 0.028382741234771514 | 0.6801256133979957 | 233.89292872 |
| ATE: SD-SLAM freiburg 1 | |||||||
|---|---|---|---|---|---|---|---|
| sequence_name | rmse | mean | median | std | min | max | sse |
| rgbd_dataset_freiburg1_desk2-groundtruth.txt | 0.171681 | 0.165224 | 0.171674 | 0.046639 | 0.033321 | 0.250823 | 17.065639 |
| rgbd_dataset_freiburg1_floor-groundtruth.txt | 0.3982463325554 | 0.166139265768752 | 0.047961755597559 | 0.361936300698957 | 0.016900295855399 | 1.94518345664361 | 169.54355115 |
| rgbd_dataset_freiburg1_360-groundtruth.txt | 0.386490484422627 | 0.36100832946858 | 0.371000997303242 | 0.138014059441575 | 0.031978899293128 | 0.648516522534314 | 107.69929897 |
| rgbd_dataset_freiburg1_rpy-groundtruth.txt | 0.028173332356165 | 0.025909035951603 | 0.023595188378316 | 0.011066097420026 | 0.00627136348811 | 0.102311778403075 | 0.49846662 |
| rgbd_dataset_freiburg1_xyz-groundtruth.txt | 0.062696784302744 | 0.062446872342138 | 0.061660441127193 | 0.005592396319064 | 0.046251918879113 | 0.08995476641068 | 2.88920177 |
| rgbd_dataset_freiburg1_desk-groundtruth.txt | 0.123545576562324 | 0.120510015489179 | 0.120974914755085 | 0.027218480025798 | 0.036184527079955 | 0.187557084643583 | 8.34913969 |
| rgbd_dataset_freiburg1_room-groundtruth.txt | 0.290971698472222 | 0.1813401797353 | 0.122264753710953 | 0.227552781844957 | 0.020909567188251 | 1.19017474767363 | 99.650151 |
As can be seen, SD-SLAM performs better in all sequences compare to rosify_difodo. There is a special case where rosify difodo seems to do it really bad, on sequence rgbd_dataset_freiburg1_floor. This is the visualization using slam-testbed tool on the f1_floor sequnce.
| Rosify_Difodo f1_floor | SD-SLAM f1_floor |
|---|---|
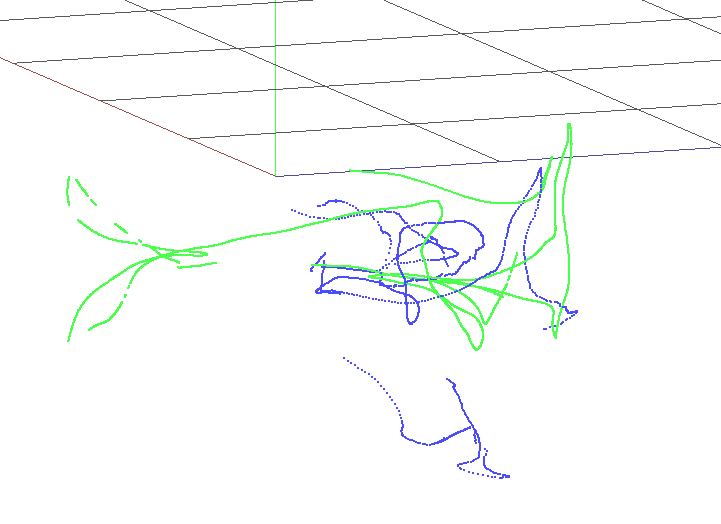 |
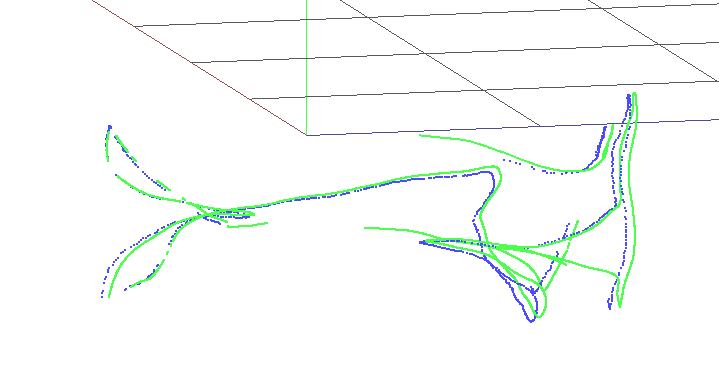 |
Since this sequence is pointing all time at the floor, we have an enviromente with low texture on it and that is the main reason why the performance of rosify_difodo in this case seems to be that bad. Also the rosbag sequence has some cuts (probably due to a problem with the file sequence when it was recorder) so there are a few frames skipped towards the end of the video that also makes rosify difodo to fail. SD-SLAM in this cases dont fail so much since it has the capability of an SLAM algorithm that allows him to relocalize in most of the cases.
From this metrics we conclude that SD-SLAM performs better in all the cases.
Freiburg 3 sequences
| ATE: Rosify Difodo freiburg 3 | |||||||
|---|---|---|---|---|---|---|---|
| sequence_name | rmse | mean | median | std | min | max | sse |
| rgbd_dataset_freiburg3_walking_halfsphere-groundtruth.txt | 3.127924554627861 | 3.038564256442317 | 3.0942872668839265 | 0.7423201997218257 | 0.03785155214783138 | 4.299877701749202 | 10028.509819930001 |
| rgbd_dataset_freiburg3_nostructure_texture_far-groundtruth.txt | 1.6508766236356571 | 1.3815921953093704 | 1.4423013875600927 | 0.9036573644512068 | 0.04349505719044416 | 2.8107884587780703 | 1226.42713191 |
| rgbd_dataset_freiburg3_walking_static-groundtruth.txt | 1.4544618033709862 | 1.1461213975180653 | 0.5440749207905468 | 0.8954690835625864 | 0.040135395849549026 | 2.8554390030256296 | 1518.8996607000001 |
| rgbd_dataset_freiburg3_nostructure_notexture_near_withloop-groundtruth.txt | 2.523445137595058 | 2.2225861913175256 | 2.099810079507192 | 1.1949418331520565 | 0.027287542945454178 | 4.285643747443317 | 6985.449572609999 |
| rgbd_dataset_freiburg3_sitting_xyz-groundtruth.txt | 1.379200327295399 | 1.1871794737123817 | 1.3223042350382155 | 0.7019960398803743 | 0.04634403953045087 | 2.507426304001774 | 2333.9914770299997 |
| rgbd_dataset_freiburg3_structure_notexture_near-groundtruth.txt | 0.2475989037621381 | 0.19206083913271546 | 0.148696555620559 | 0.15626212342039164 | 0.006195159400693396 | 0.6053854061670136 | 64.61569887 |
| rgbd_dataset_freiburg3_long_office_household-groundtruth.txt | 0.9000034454921396 | 0.6838115581922141 | 0.3845090896194783 | 0.5851563507136011 | 0.007308898685848572 | 1.9110632040830045 | 2027.4455233499998 |
| rgbd_dataset_freiburg3_structure_texture_far-groundtruth.txt | 0.7341830189114689 | 0.6620370886288545 | 0.7307891761650551 | 0.31738241686928514 | 0.003401470270338909 | 1.077531354532201 | 491.05150648999995 |
| rgbd_dataset_freiburg3_sitting_rpy-groundtruth.txt | 2.119234935191984 | 2.0228722972722615 | 2.277127159383068 | 0.631778742493458 | 0.045237373929086576 | 2.5875250066424478 | 3588.43421172 |
| rgbd_dataset_freiburg3_sitting_static-groundtruth.txt | 0.28225323647651285 | 0.23857610021717482 | 0.18397208699419465 | 0.1508255081431216 | 0.0394171282566347 | 0.5477462916350964 | 54.33281864 |
| rgbd_dataset_freiburg3_structure_texture_near-groundtruth.txt | 0.3622751995740463 | 0.32150006223790956 | 0.26867994829031494 | 0.16697613664064512 | 0.009593226777263196 | 0.7236220975067026 | 139.11791943999998 |
| rgbd_dataset_freiburg3_walking_rpy-groundtruth.txt | 1.8357329237983508 | 1.5897241756503455 | 1.5337489405833935 | 0.9179827955196493 | 0.04672076626083959 | 3.5109668568643597 | 2931.8263697400002 |
| rgbd_dataset_freiburg3_nostructure_texture_near_withloop-groundtruth.txt | 2.4571612872409334 | 2.2807206186364484 | 2.165508265973603 | 0.9143057755707861 | 0.03737285110879301 | 3.474077052973926 | 9919.84513486 |
| rgbd_dataset_freiburg3_sitting_halfsphere-groundtruth.txt | 0.48470849992326775 | 0.4172286528861979 | 0.455472820264832 | 0.24670342743592555 | 0.041706114659603534 | 0.9978358482235441 | 253.03288930000002 |
| rgbd_dataset_freiburg3_walking_xyz-groundtruth.txt | 3.073620672389294 | 2.5129130830178177 | 2.3198924587144125 | 1.7698621061926554 | 0.05597463711360713 | 5.934649521243862 | 7812.78811921 |
| rgbd_dataset_freiburg3_nostructure_notexture_far-groundtruth.txt | 1.9231111863962236 | 1.6896934207020713 | 1.7790875179210475 | 0.9183097403808936 | 0.04488886276126859 | 3.101961716398189 | 1679.0539124 |
| rgbd_dataset_freiburg3_structure_notexture_far-groundtruth.txt | 0.23194002589710105 | 0.1943633084806683 | 0.17585602065326053 | 0.12656650397951466 | 0.014439182802360994 | 0.42580400420850917 | 42.55277491 |
| ATE: SD-SLAM freiburg 3 | |||||||
|---|---|---|---|---|---|---|---|
| sequence_name | rmse | mean | median | std | min | max | sse |
| rgbd_dataset_freiburg3_walking_halfsphere-groundtruth.txt | 0.49535373929853105 | 0.4553007329607464 | 0.5037697005697832 | 0.19513218494765056 | 0.027993927913031454 | 0.823883857106085 | 225.25455022 |
| rgbd_dataset_freiburg3_nostructure_texture_far-groundtruth.txt | 1.417193362139913 | 0.5861165239307159 | 0.07381456495841449 | 1.290311763128898 | 0.0020784609690826373 | 3.91645190829659 | 825.46761756 |
| rgbd_dataset_freiburg3_walking_static-groundtruth.txt | 0.047673151886864225 | 0.040201770751377194 | 0.03589045831761439 | 0.0256231738721365 | 0.009148770409186052 | 0.2749760898696467 | 1.4272740700000002 |
| rgbd_dataset_freiburg3_nostructure_notexture_near_withloop-groundtruth.txt | 2.209193148457564 | 1.951563150642346 | 2.3724186603548705 | 1.035343245617976 | 0.010768008172359527 | 3.3740151066644617 | 5266.0965822 |
| rgbd_dataset_freiburg3_sitting_xyz-groundtruth.txt | 0.044595105746728786 | 0.044191966170789514 | 0.04400812419976659 | 0.00598277381502241 | 0.02196132965009151 | 0.0653738479822016 | 2.1517987800000005 |
| rgbd_dataset_freiburg3_structure_notexture_near-groundtruth.txt | 2.0055327666395337 | 1.7475283309053462 | 1.6270156391381123 | 0.9840256148840834 | 0.009403722667114407 | 3.359556499003998 | 4219.247600289999 |
| rgbd_dataset_freiburg3_long_office_household-groundtruth.txt | 0.04969134528462328 | 0.049002083815100385 | 0.04902351472956422 | 0.008247761997871216 | 0.029305801473428456 | 0.08715096098150604 | 5.4520593900000005 |
| rgbd_dataset_freiburg3_structure_texture_far-groundtruth.txt | 0.0437790678254005 | 0.042739350269403335 | 0.041320333977353 | 0.009484446120373594 | 0.023095021108455446 | 0.06853553822652886 | 1.4700374000000003 |
| rgbd_dataset_freiburg3_sitting_rpy-groundtruth.txt | 0.048230236617063466 | 0.046364098252378944 | 0.04433035077686612 | 0.013286313159852937 | 0.009120307012376204 | 0.14844938531364818 | 1.686462899999999 |
| rgbd_dataset_freiburg3_sitting_static-groundtruth.txt | 0.05321483855941207 | 0.05299509243851639 | 0.05258084249551545 | 0.004831068239778241 | 0.04199345187049997 | 0.06688437784714747 | 1.7160823399999998 |
| rgbd_dataset_freiburg3_structure_texture_near-groundtruth.txt | 0.046698864071617406 | 0.04612439169548674 | 0.045660814489485946 | 0.007302355530971717 | 0.026449196585151633 | 0.06855953617112652 | 2.0324906 |
| rgbd_dataset_freiburg3_walking_rpy-groundtruth.txt | 0.7251566200072731 | 0.576554308416373 | 0.4659282455486036 | 0.4398150213292977 | 0.03509344668168125 | 1.9085011605969748 | 423.31095945 |
| rgbd_dataset_freiburg3_nostructure_texture_near_withloop-groundtruth.txt | 0.06657011390633324 | 0.06544519717549836 | 0.06361142351605716 | 0.012186313312988648 | 0.03789485453198092 | 0.10033214838724436 | 6.088991009999999 |
| rgbd_dataset_freiburg3_sitting_halfsphere-groundtruth.txt | 0.05072051609139362 | 0.04898224447767757 | 0.04658234569878228 | 0.013164743769110906 | 0.023021946051539496 | 0.09244317173269213 | 2.4953936299999997 |
| rgbd_dataset_freiburg3_walking_xyz-groundtruth.txt | 0.40001104723626224 | 0.37342579391369307 | 0.3358192246773971 | 0.14339461060646333 | 0.035992915969673786 | 0.976198489037962 | 118.72655773000001 |
| rgbd_dataset_freiburg3_nostructure_notexture_far-groundtruth.txt | 1.7486785481708642 | 1.514286051209603 | 1.504421912230741 | 0.874536688735807 | 0.047425942267919165 | 2.9872047703496993 | 1372.9866225099997 |
| rgbd_dataset_freiburg3_structure_notexture_far-groundtruth.txt | 2.6243429265067877 | 2.310724596674208 | 2.5927306609056022 | 1.2440770210204146 | 0.02532093995095747 | 3.860584747159425 | 5433.98170297 |
In this case once again, SD-SLAM seems to be the big winner in almost all the cases. But this time there are some sequences were rosify difodo performs better than SD-SLAM. Lets take a deep look into those sequences that has less error in rosify difodo and also look for sequences were the error is too big either for rosify difodo or SD-SLAM.
No texture cases
Here the cases were no texture is provided but there is structure are compare. The expected is that in this case rosifu_difodo does better than SD-SLAM since its based on depth only while SD-SLAM is based on visual information (texture).
rgbd_dataset_freiburg3_structure_notexture_near
In this case we see how difodo performs way better than SD-SLAM. This is the expected behaviour since in this case there is no texture for SD-SLAM to work. In fact SD-SLAM didnt even initialized so no odometry at all was provided. We see that with enough texture rosify_difodo gets a 0.2475 RMSE on ATE on this sequence with a distance of 0.36 seconds.
Rosify_difodo with slam-testbed
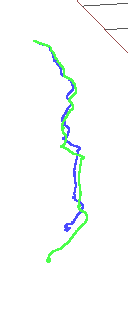
rgbd_dataset_freiburg3_structure_notexture_far
Once again another case where there is no texture and a lot of structure. As in the case before the results are similar. While rosify_difodo does pretty decent and get a 0.2319 RMSE on ATE, SD-SLAM does get a 2.6243 RMSE on ATE.
Rosify_difodo with slam-testbed
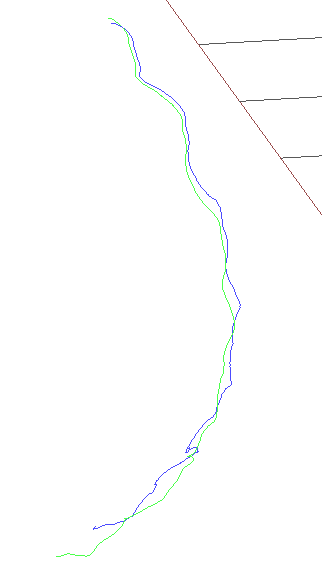
In this section we can conclude that the expected results have been obtained.
No structure cases
In this scenario the inverse results are expected. We expect rosify_difodo to perform badly while SD-SLAM should do it good.
rgbd_dataset_freiburg3_nostructure_texture_near_withloop
In this case we see how rosify_difodo struggles and gets a really bad ATE of more than 2.4 meters. This is expected since in this sequence there is no structure at all so is the opposite case to the previous ones. In this case SD-SLAM performs quite good with only 0.06m of ATE error.
| Rosify_Difodo f3_nostructure_texture_near_withloop | SD-SLAM f3_nostructure_texture_near_withloop |
|---|---|
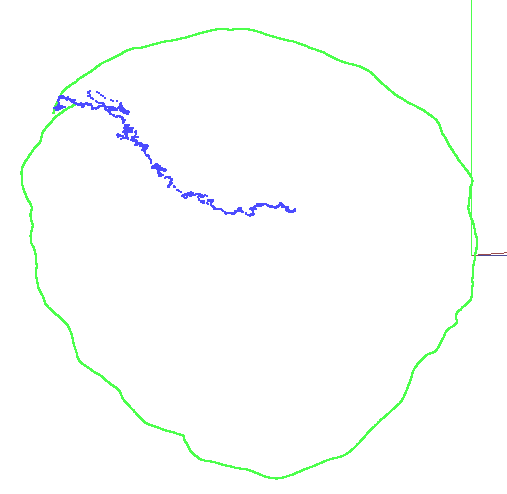 |
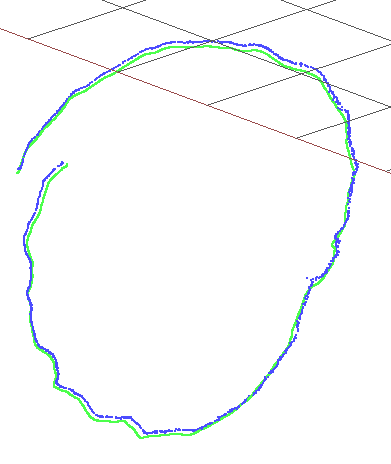 |
rgbd_dataset_freiburg3_nostructure_texture_far
In this case both rosify_difodo and SD-SLAM perform bad whit a high error of 1.65m RMSE for rosify_difodo and 1.41m for SD-SLAM, although the mean for SD-SLAM is much less 0.58m while in rosify_difodo is more or less the same. If we see the median of SD-SLAM this is only 0.07m, so here we see a case that the algorithm has perform really good up to a certain point where it got lost or something. So for SD-SLAM there are some outliers that are adding a lot of error. After reviewing the results from SD-SLAM we can see that the high error is due to the last 2-3 seconds of video where the camera view goes out of the texture zone and the only thing that can be seen is the floor and a cable, see next image:

In those last seconds SD-SLAM gets lost due to the absence of texture and because of that we got the high RMSE and mean but a low median value.
Still if we look at the trajectories generated with the tool slam-testbed we can see that the results are good for SD-SLAM, not so good for rosify_difodo:
| Rosify_Difodo f3_nostructure_texture_far | SD-SLAM f3_nostructure_texture_far |
|---|---|
 |
 |
Other cases
rgbd_dataset_freiburg3_walking_halfsphere This is a very challenging sequence since we have people walking in front of the camera which makes rosify_difodo and also SD-SLAM fail since both algorithms cannot handle non static environments due to their nature. Rosify Difodo since is an odometry method suffers much more than SD-SLAM which can relocalize more effectively in this case since the camera doesnt change of position almost.
rgbd_dataset_freiburg3_walking_xyz The same as with the previous sequence. All “walking” sequences have moving objects in the scene which make rosify_difodo and also SD-SLAM fail.
Conclusions
SD-SLAM outperforms rosify_difodo in most of the situations. While rosify_difodo moves in the order of 0.2-0.3m of ATE for an appropiate environment, SD-SLAM has a mean of 0.05m for appropiate environments. This means that SD-SLAM is 4-6 times more accurate when it comes to accuracy on the pose estimation task.
It seems that the visual information is much richer that the structure information at this moment and also with the current state of the art algorithms.
Also rosify_difodo is only odometry while SD-SLAM is a full SLAM algorithm which gives SD-SLAM some advantages like reducing the drift with relocalizations.
Even though SD-SLAM seems better, there are cases where SD-SLAM fails to give a good pose estimation. Not just a good pose but even to initialize. This scenarios are the ones with low texture information due to the environment or could it be for example due to lightning conditions (dark places) or even in the completely dark. In this cases mention, rosify_difodo has a lot to say, since due to its nature of using only depth information, its not affected by the lightning conditions, being able to work on completely dark environments and also when texture is not enough.
With this advantages and disadvantages in mind, we are going to try to merge this two algorithms in order to create a better version of SD-SLAM new one that can outperform the current SD-SLAM and work in situations where SD-SLAM cannot.